Research Highlights
NCMIR researchers help biologists reveal greater structural detail.
November 2006
In step with its mission of advancing the analysis of 3D structures in neurobiology and cell biology, a team of NCMIR researchers is helping biologists to take advantage of large-field electron microscope imaging developments for improving the resolution of 3D structures at the scale of proteins. Albert Lawrence and NCMIR colleagues from the Center for Research in Biological Systems (CRBS) recently published a technique to increase the resolution of electron microscope-derived tomograms. A major benefit of the mathematically sophisticated model is the ease of adapting the code for parallel computation. This work was published in the May issue of the Journal of Structural Biology.
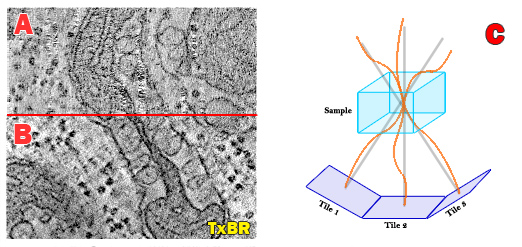
The new transform-based back projection (TxBR) method complements other NCMIR-led efforts focused on developing improved large-field detectors for electron microscopy. Until now, large-field 3D reconstructions from a standard tilt series of individual electron micrographs assumed a linear back projection geometry that ignored the curvilinear trajectories of electrons passing through the strong magnetic fields of the electron microscope that distort images.
NCMIR’s Albert Lawrence applied his passion for topology and global geometry to develop a global transform to compute the curvilinear trajectories of electrons and eliminate some of the distortions inherent in large-field electron microscope volume tomograms. The new code is more rigorous and compact—only a fraction of the size of the code used in comparable algorithms. The mathematical point of view permits easy extension to more complex projection models.
Accommodating the complexity of the electron microscope as an optical instrument—without having to untangle each separate phenomenon’s impact on image distortion—is unique to the TxBR technique. In effect, optical distortion, beam effects, sample warping and positioning errors are treated in a lumped parameter model represented by simple polynomial functions. These effects are almost impossible to untangle so a lumped parameter model calculation from the actual data is the most effective approach. By compensating for the inherent geometric nonlinearities of the electrons path through the microscope, this algorithm helps reconstruct the smaller image scales representing sub-cellular structures, ultimately providing crisper resolution.
Public release of the TxBR MATLAB M-files (The MathWorks) are planned to coincide with the 4th Intl. Congress on Electron Tomography, November 5-8, 2006.
Citation: Lawrence A, Bouwer JC, Perkins G, Ellisman MH. (2006) Transform-based backprojection for volume reconstruction of large format electron microscope tilt series. J Struct Biol. 154(2):144-67