Research Highlights
PRIME Directive: New Chemical Fluorophore Designed to Tag Cell Proteins for Imaging
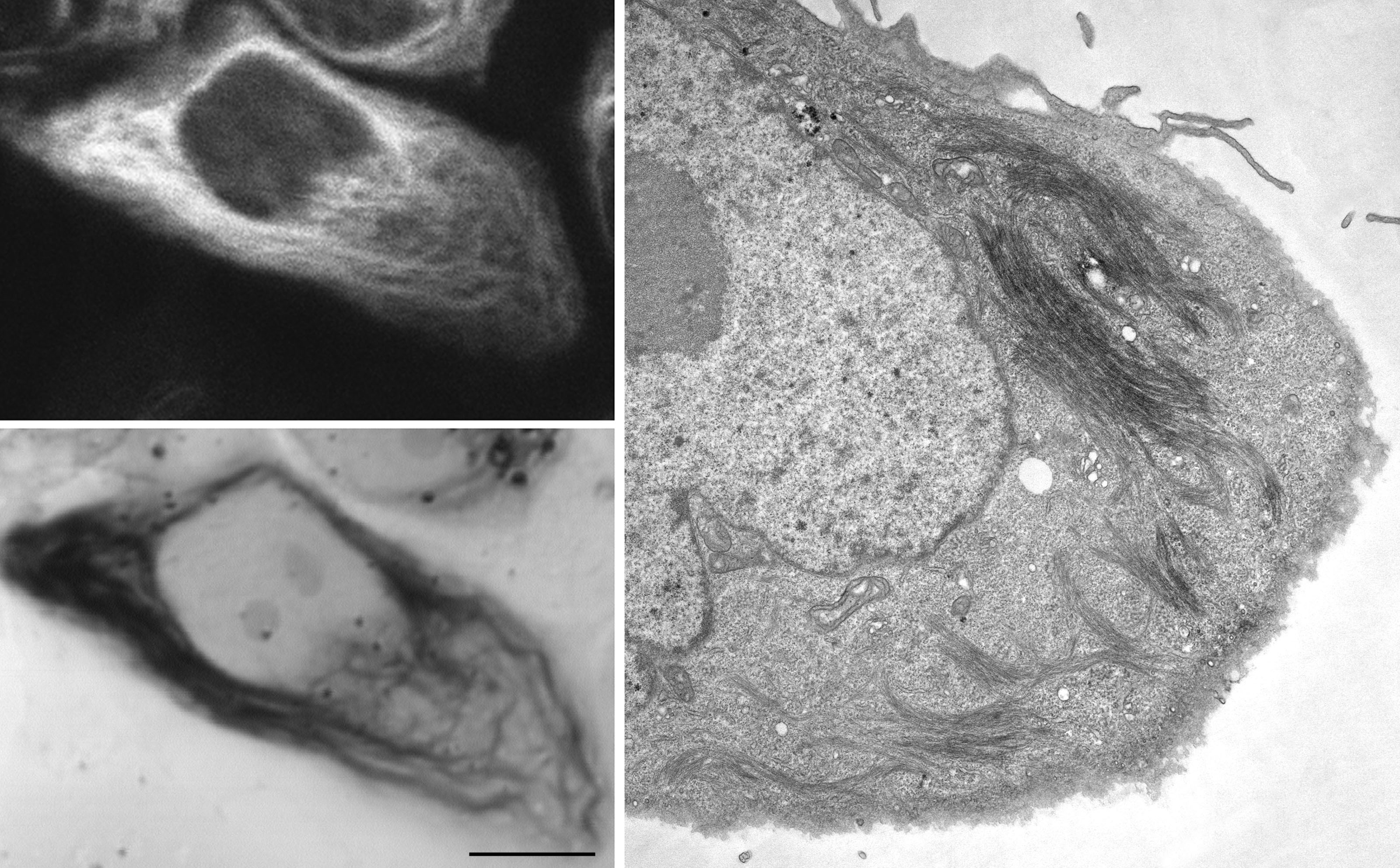
Figure Caption: Correlated light and electron microscopic imaging of site-specific resorufin ligase labeled vimentin filaments in cultured CV-1 cells. Left panels: confocal fluorescence image (top) and transmitted light image following fluorescence photooxidation of DAB (bottom). Right panel: TEM image of the same cell.
January 2015 La Jolla -- Fluorescent proteins are used commonly to tag components of interest in living cells for subsequent imaging. But there’s a cost: Their dim fluorescence, rapid photobleaching, and large size limit their utility, and they can disrupt protein folding and trafficking and/or impair protein function.Chemical fluorophores, by comparison, tend to be considerably smaller, brighter, and more photostable, characteristics that allow them to perform better in advanced biological imaging modalities such as single-molecule tracking and super-resolution microscopies. However, it is much more challenging to target them to specific cellular proteins inside living cells because they cannot be genetically encoded and have to target proteins post-translationally inside a complex cellular environment. In this context, a research team, representing the Howard Hughes Medical Institute, MIT, the University of Washington, UCSD, and the Max Planck Institute for Biophysical Chemistry, used the enzyme redesign capabilities of the Rosetta algorithm to create a new technology for protein labeling and imaging in living cells.
To achieve labeling specificity for chemical fluorophores comparable to that of fluorescent proteins, the team developed PRIME(PRobe Incorporation Mediated by Enzymes), which uses Escherichia coli lipoic acid ligase (LplA) to attach small molecules to a peptide tag. To make PRIME more useful for cellular protein imaging, they sought to engineer the system to target bright chemical fluorophores. The challenge they faced was that lipoic acid ligase has a small, fully enclosed substrate-binding pocket that previously was not able to accommodate large chemical structures.In this work, the collaborating interdisciplinary group demonstrated that PRIME with resorufin ligase can be used to study specific cellular proteins in an optimal spectral window while minimizing steric perturbation to those proteins, demonstrating that PRIME represents an attractive alternative to red fluorescent proteins, which are considerably larger and inappropriate for use in cases where bulky tags are not tolerated.
This study also showed that, despite the post-translational requirement of the labeling, the tagging is exceptionally specific, comparable to that obtained by genetic fusion to fluorescent proteins. In addition to conventional fluorescence microscopy, the team showed that resorufin PRIME can be used for super-resolution fluorescence imaging by techniques such as stimulated emission depletion (STED) and higher-resolution imaging by electron microscopy. STED selectively restricts emission from fluorophores to a small central spot to enhance the axial resolution of labeled structures in a particular area of a biological sample. For electron microscopy, the resorufin fluorophore can be used to photochemically polymerize DAB, thereby creating EM contrast, allowing for directly correlated superresolution LM and EM. Such multi-modality tags are rare but provide great versatility for biological applications, particularly for the increasing number of investigations using new methods for correlated light and electron microscopy. However, additional work remains as resorufin’s photophysical properties are not optimal for long-term fluorescence imaging, nor is resorufin highly efficient in the fluorescence photooxidation used for electron microscopy.
From the standpoint of computational design, this work is notable in two key respects. First, the redesign from lipoic acid recognition to resorufin recognition required a larger change in size, shape, and location of the binding pocket than in previous redesigns. Second, although the team explored approximately 109 LplA variants in silico, they found an unusually strong convergence toward a single sequence, and the top-ranked conformer of the sequence closely resembled the crystal structure. In summary, this work illustrates the power of computational design to develop enzymes with practical utility for cell biology.
This work was funded by NIH grants DP1 OD003961 and R01 GM072670, NIH Grant P41 GM103412, and the American Chemical Society. Drennan is a Howard Hughes Medical Institute (HHMI) Investigator. Nivón is supported by a National Science Foundation Minority Postdoctoral Fellowship. Ye has support from the Lord Foundation and the HHMI-Massachusetts Institute of Technology Summer Research Program in Chemical Biology.
Citation: Liu, Daniel S. Lucas G. Nivón, Florian Richter, Peter J. Goldman, Thomas J. Deerinck, Jennifer Z. Yao, Douglas Richardson, William S. Phipps, Anne Z. Ye, Mark H. Ellisman, Catherine L. Drennan, David Baker, and Alice Y. Ting, Computational design of a red fluorophore ligase for site-specific protein labeling in living cells, PNAS, published online October 13, 2014, pp. E4551-E4559. This article contains supporting information online at http://www.pnas.org/content/suppl/2014/10/08/1404736111.DCSupplemental/pnas.1404736111.sapp.pdf.
Link to Article in PubMed Central