Research Highlights
Epik! A New Scientific Workflow for Electron Tomography
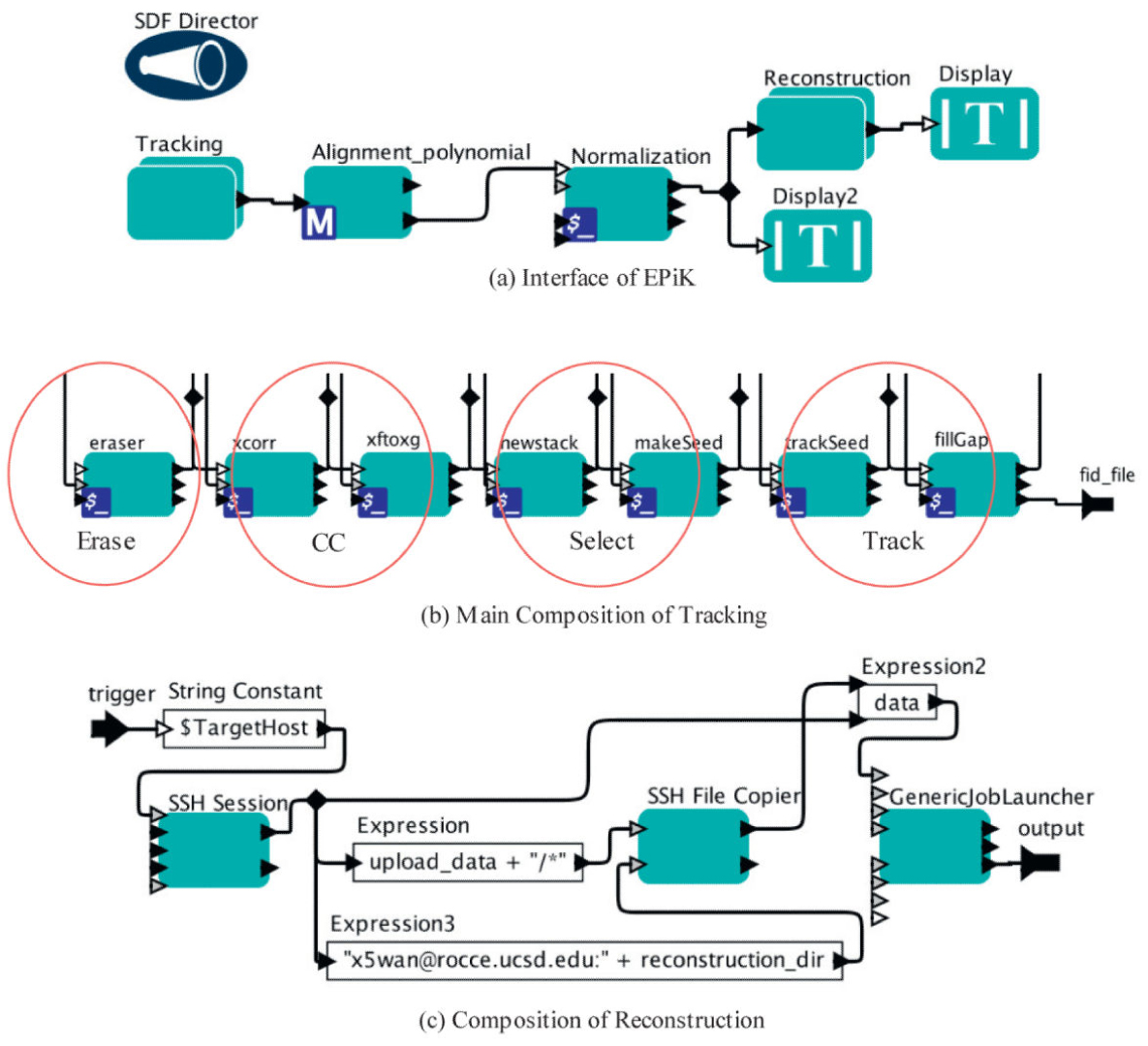
Figure Caption: EPiK Workflow. (a) The main interface of EPiK. It includes three main parts: tracking, alignment and reconstruction. Normalization is a pre-reconstruction step to insure that the grey-scale statistics are correct. (b) Main composition of tracking. IMOD is used for coarse tracking, and TxBR is used for fine tracking. All of the steps are integrated as a composite actor in EPiK. (c) Composition of reconstruction. There are parallel computings in this step. Multiple nodes in a cluster are used for large data sets.
February 2015 La Jolla -- Computational researchers, regardless of their disciplines, need software tools that save time, optimize and scale up computations, produce results faster, create an extensible platform, foster collaborations, effectively communicate the underlying science, and enable others to replicate the results. Some of the most effective tools are based on scientific workflows. A workflow is a software application that solves a scientific problem. It’s composed of computational steps and data manipulation tools that can scale up to run on high-performance computers, distributed environments, and commercial cloud systems. Workflows are used in all stages of the data lifecycle: generation and acquisition, analysis, comparison, publication, and archiving. Because they are flexible in their application, they provide a common language that fosters scientific collaborations. And they serve as a practical tool to explain scientific methodology to colleagues and elucidate articles in peer-reviewed journals.
Many of today’s scientific applications are data- and information-driven, and structured as pipelines or workflows with a large number of distinct computations. Scientists working in many computation-intensive domains, ranging from large-scale astronomy to small-scale bioinformatics, have adopted workflows successfully. In general, workflow applications put together data sets from one or more data sources, transform the data into a format amenable for processing, analyze the data, and store the data and results in a repository that scientists can access. Many of the steps in data access and processing are distributed across different execution sites, requiring data to be moved across a network for subsequent processing by the next step(s) in the workflow. Thus, scientific workflows are graphs of analytical steps that may involve, e.g., database access and querying steps, data analysis and mining steps, and many other steps including computation-intensive jobs on high-performance cluster computers.
Kepler, one of the mature workflow management systems, provides a visual interface that can be used to define and build the processing required in a workflow, and raise the level of abstraction employed in a workflow application solution. An ideal area to apply workflows is in the field of electron tomography (ET), a powerful technique that enables 3D imaging of cellular ultrastructure. Large-field, high-resolution ET facilitates visualizing and understanding global structures, such as the cell nucleus, extended neural processes, or even whole cells on scales approaching molecular resolution.
ET images can run to a terabyte in size. So this field depends on mass data processing integrated with complex algorithms using high-performance computers to achieve results. In this context, researchers from UC San Diego and the Chinese Academy of Sciences published a new scientific workflow tailored to the needs of ET. It’s called EPiK, for Electron Tomography Programs in Kepler. This workflow embeds various ET processes like tracking (using IMOD), alignment (TxBR), and reconstruction. IMOD is a set of image processing, modeling, and display programs, developed at the University of Colorado, used for 3D reconstruction of the images. The package contains tools for assembling and aligning data, viewing 3D data from any orientation, and modeling and displaying the image files. TxBR, developed at UCSD, is an advanced ET code developed to compensate for curvilinear electron trajectories and sample warping to provide better alignment and reconstruction quality. It also enables 3D reconstruction from various schemes of data acquisition including tilt series. A tilt series is a sequence of images taken of a tissue sample as the platform on which it sits is tilted around one or more axes in regular (such as one-degree) increments; these 2D images can then be reconstructed into a 3D volume.
High-resolution tomography of complex biological specimens leads to the requirement for large reconstructed files. Reconstruction requires extensive use of computational resources and considerable processing time. In response to this requirement, TxBR has been adapted for various parallel computers, computer clusters, and processors with multiple graphical processor unit (GPU) boards. By using fast recursion algorithms and other parallel processing with GPU for algorithms such as backprojection, TxBR can achieve significant speedups on relatively inexpensive hardware comprised of commercial off-the-shelf components. EPiK provides users the flexibility to choose among available computational resources based on the size of their data sets.
Besides providing useful technical tools, EPiK, significantly, also facilitates scientific collaboration. There are many ET research groups in different parts in the world, and scientists at each site typically develop and use their own codes and resources to conduct their research. But contrary to published claims, it is difficult or impossible to make fair comparisons of different algorithms on common data sets. EPiK addresses this problem because it can be used to integrate many software packages and make it possible to compare and cooperate on methods proposed by different research groups.
The UCSD scientists tested the 3D reconstruction process at the National Center for Microscopy and Imaging Research at UCSD using EPiK on ET data taken from the electric organ of an eel. This organ can generate a high-voltage pulse for self defense. Reconstruction of the organ’s structure can help biologists understand its physiological functions.
This work was supported by grants from an International Community Foundation, San Diego, the National Institutes of Health (GM103412, GM103426), the National Science Foundation (NSF DBI-1062565), and the National Science Foundation for China (61232001, 61202210, and 60921002).
Citation: Chen, Ruijuan, Xiaohua Wan, Ilkay Altintas, Jianwu Wang, Daniel Crawl, Sébastien Phan, Albert Lawrence, and Mark Ellisman, EPiK: A Workflow for Electron Tomography in Kepler, Procedia Computer Science, Vol. 29, 2014, pp. 2295-2305, doi: 10.1016/j.procs.2014.05.214.
Link to Article in PubMed Central