Research Highlights
X-ray Microscopy Increases Accuracy and Efficiency of SBEM Imaging for Correlated Light/Electron Microscopy of Biological Specimens
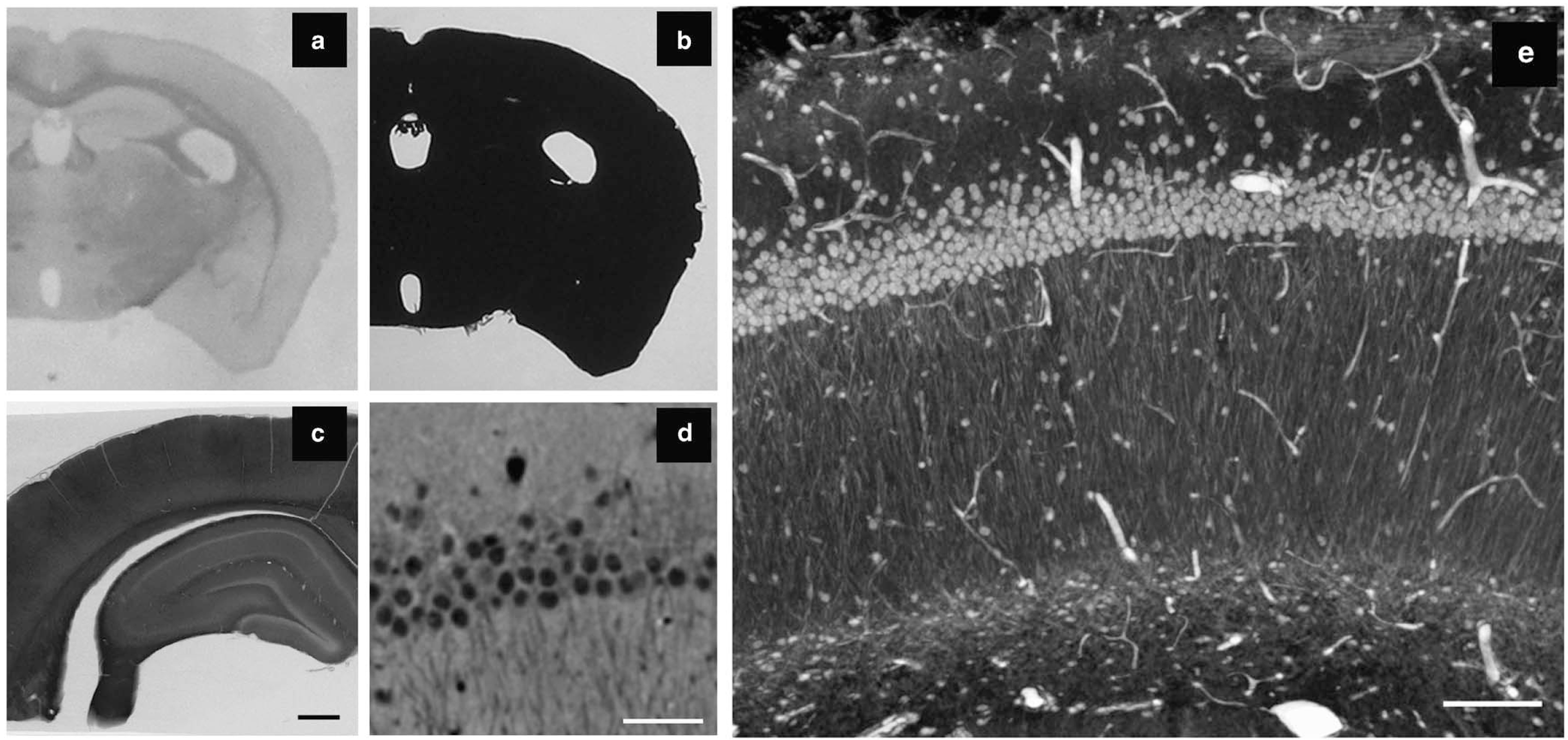
Figure Caption: X-ray microscopy (XRM) allows for high-resolution imaging of tissue stained for serial block-face scanning electron microscopy (SBEM). (a, b) While it is possible to easily identify anatomical features within a brain slice using light microscopy, the tissue is completely opaque to light following staining for SBEM. (c) A two-dimensional (2D) projection image with the XRM using a 4× objective and 40 kV reveals details within the SBEM-stained tissue slice, including cell layers, vasculature, and white matter tracts. (d) A virtual 2D slice from a XRM volume allows for the discrimination of individual cells, dendrites, dark neurons, and nucleoli. The volume was collected with a 20× objective using a 180° sample rotation, 0.1° tilt increments, and 15 sec exposures (~7.5 hours total). (e) A 3D ray trace projection image of the same volume. Scale bar: (c) 500 μm, (d) 50μm, and (e) 100 μm.
February 2015 La Jolla -- The recently developed 3D method of serial block-face scanning electron microscopy (SBEM) is rapidly establishing itself as a powerful imaging approach in the biological sciences research community. This approach is proving of great value for 3D visualization of nervous system ultrastructure, particularly when details of synaptic and other subcellular elements must be located and quantified. It is also important for constructing connectomic models of regional brain circuits. In addition, SBEM is increasingly used to perform nanohistology on a wide variety of tissue systems, including the lung, liver, tendons, kidney, and cell cultures.
SBEM imaging entails an iterative process of imaging a specimen blockface followed by removal of a thin layer of epoxy-embedded tissue, then imaging again. As such, SBEM is a destructive technique, allowing only a single opportunity to collect images of an object of interest. In addition, typical experiments can require days or weeks of automated SBEM machine processing time. This method requires staining of biological specimens with heavy metals to enable sufficient back-scatter electron signal and render specimens sufficiently conductive to control charging artifacts. But these staining protocols render specimens light-opaque. As a result, their use makes it much more problematic to track and identify regions of interest (ROIs) in the SBEM imaging process than for a typical thin-section transmission electron microscopy correlative light and electron microscopy study.
Further complicating correlative work is the fact that once a sample is placed in the SEM for imaging, only the freshly prepared blockface is visible. This can create uncertainty when deciding where to collect data from the sample surface to reach a ROI deep within the sample. In practice it is common to collect a larger volume than necessary to ensure that the ROI is captured. This practice results in wasted microscope time and/or a sacrifice in image quality owing to lower-resolution scans.So any methods that can improve the precision and efficiency of SBEM imaging would greatly enhance the power of the technique and availability of rare imaging resources.
In this context, a research team of NCMIR scientists, Carl Zeiss X-ray Microscopy, Inc., and the Salk Institute for Biological Studies developed a strategy employing X-ray microscopy (XRM) as part of an improved SBEM correlative imaging workflow to track ROIs and increase efficiency of typical SBEM projects. XRM is a tomographic technique that uses X-ray illumination to reveal the interiors of optically opaque samples in a nondestructive manner. Modern laboratory-based XRM is capable of producing 3D tomographic reconstructions routinely with submicron resolution. The team found that XRM reveals an impressive level of detail in tissue heavily stained for SBEM imaging, enabling identification of tissue landmarks that can be used subsequently to guide SEM data collection. Furthermore, specific labeling of individual cells using diaminobenzidine is detectable in XRM volumes. The team demonstrated that tungsten carbide or nanophosphor particles can be used as fiducial markers to increase the precision and efficiency of tracking ROIs within an SBEM sample.
In summary, the team demonstrated the ability to accurately target subsurface volumes of interest within optically opaque biological specimens for 3D SBEM imaging using XRM as a bridging imaging modality. Accuracy is enhanced by acquiring 3D XRM volumes of specimens with their coordinate space accurately marked using fiducial markers. These techniques provide sufficient accuracy to enable targeting of single cells or even subcellular domains (e.g., a particular subregion of a dendritic field) that have been previously imaged at lower resolution using light microscopy. In addition, mapping an ROI with XRM greatly increases the ability to use SBEM to target structures previously labeled with EM dense stains using approaches such as photooxidation.
In addition to increasing the efficiency of SBEM imaging, the integration of XRM in the correlative imaging workflow offers the potential to correct for many of the spatial distortions to biological samples that occur during specimen preparation for EM. Since the ultimate goal of many SBEM studies is to reconstruct cells from large 3D volumes, the ability to correct for shrinkage and compression artifacts inherent in EM specimen preparation procedures could increase the fidelity of reconstructions.
This work was supported by an NIH National Institute of General Medical Sciences grant (5P41GM103426).
Citation: Bushong, Eric A., Donald D. Johnson, Jr., Keun-Young Kim, Masako Terada, Megumi Hatori, Steven T. Peltier, Satchidananda Panda, Arno Merkle, and Mark H. Ellisman, X-ray Microscopy as an Approach to Increasing Accuracy and Efficiency of Serial Block-Face Imaging for Correlated Light and Electron Microscopy of Biological Specimens, Microscopy and Microanalysis, pp. 1-8, doi:10.1017/S1431927614013579.